Use cases: Cancer
The Cancer Use Case has been the secondary use case to test the 1+MG Infrastructure. Although a work still in progress it has been acknowledged that the architecture largely supports the Cancer Use Case in its current format.
The expected outcomes from the 1+MG Inititaive for this use case include
- Aligning cancer centres over signatory countries
- Agree on a minimal oncological dataset and its standards
- Survey the current genomic approaches in clinical practice
- Establishing a Proof of Concept on data exchange (with WP4)
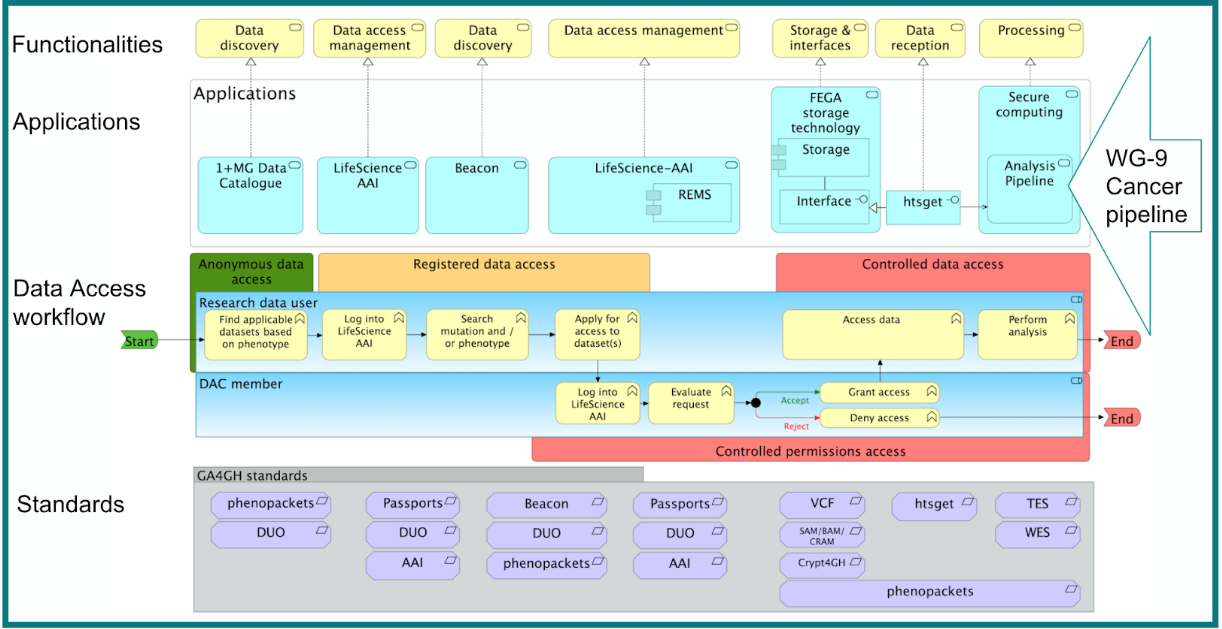
B1MG Cancer Proof of Concept: In progress
Informational
- The objective of the Cancer Proof of Concept follows the same principles as that of the Rare Disease Use Case i.e., define a set of standards, services and components that can support the five 1+MG functionalities and demonstrate these in action - using two actors: Data User and Data Access Committee (DAC).
The scenario was placed in a healthcare context where a patient receives a BRAF-positive metastic melanoma diagnosis via a standard molecular test, has a BRAF biomarker, and starts standard therapy. After the therapy the patient returns and it is found out that the tumour no longer responds to therapy. Tumour sample is then taken, sequenced, and a mutation is found in the gene encoding PTEN protein, preventing the therapy to work.
Driving Questions for the Infrastructure:
- Was the aquired PTEN resistance mutation novel or previously described in patients with BRAF V600?
- Are different acquired PTEN mutations identified in patients with BRAF V600E and not responding to therapy?
- What treatment was used for other patients?
- Proof of Concept “starting point” for Cancer utilised a number of existing GA4GH Standards
- Beacon
- DUO
- SAM,BAM, CRAM
- VCF
- Crypt4gh
- Htsget,
- Phenopackets
- Passports
- TES & WES
- And other existing services ( FEGA & Life Science AAI).
1+MG Minimal dataset for cancer
Required
- paper in progress
- For a real impact on healthcare, precision cancer medicine requires accessibility and interoperability of clinical and genomic data across centres and countries. Due to the heterogeneous digitization in Europe and worldwide, the definition of models for standardised data collection and usability becomes mandatory if countries want to work together on this mission. The European Union’s 1+Million Genomes (1+MG) initiative, supported by the Horizon 2020 Beyond 1 Million Genome project, aims at outlining data models, guidance, best practices, and technical infrastructures for transnational access to sequenced genomes, including cancer genomes. Within the framework of the cancer-focused Working Group 9 (WG9), we developed the 1+MG-Minimal Dataset for Cancer (1+MG-MDC)–a data model encompassing 140 items and organized in eight conceptual domains, for the collection of cancer-related clinical information and genomics metadata. The 1+MG-MDC, which results from a multidisciplinary effort, leverages pre-existing models and emphasizes the annotation and traceability of multiple aspects relevant to the complex longitudinal path of the cancer disease and its treatment. We strived to make 1+MG-MDC easy to adopt, yet comprehensive, addressing the needs of both clinicians and researchers. It will be periodically revisited and updated by the 1+MG WG9 to ensure it remains fit for purpose. We propose it as a model to create homogeneous databases, which would, in turn, guide discussions on clinical and genomic features with prognostic or therapeutic value and foster real-world data research.
We are still working on the content for this page. If you are interested in adding to the page, then:
This is a community-driven website, so contributions are welcome! You will, of course, be listed as a contributor on the page.